NSIDC Ice Age Exploration
Historical Beaufort Sea conditions with Dask/Xarray
Checking out a really neat historical sea ice age dataset from NSIDC: https://nsidc.org/data/nsidc-0611
from io import BytesIO
from netrc import netrc
from pathlib import Path
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import requests
import xarray as xr
from matplotlib.colors import ListedColormap
We need to get the data first (requires a NASA EarthData account!)
local_nc = Path('./iceage_nsidc.nc')
if local_nc.exists():
ds = xr.open_dataset(
local_nc,
chunks={'time': 200, 'x': 722, 'y': 722} # try to have ~100MB chunks
)
else:
resp = input(
'Local dataset does not exist - confirm you ' \
'want to download from NSIDC? (Approx bandwidth usage: 1GB)'
)
if resp.lower() == 'y':
base_url = 'https://daacdata.apps.nsidc.org/pub/DATASETS/' \
'nsidc0611_seaice_age_v4/data/' \
'iceage_nh_12.5km_{year}0101_{year}1231_v4.1.nc'
urls = [
base_url.format(year=y)
for y in range(1984,2021) # 2021 is still preliminary
]
user, _, password = netrc().authenticators('urs.earthdata.nasa.gov')
with requests.Session() as sesh:
sesh.auth = (user, password)
ds = xr.open_mfdataset(
[
BytesIO(sesh.get(url).content)
for url in urls
],
data_vars=['age_of_sea_ice']
)
else:
raise FileNotFoundError(
'Local dataset not found user decided not to download'
)
ds
ds['age_of_sea_ice']
print('Approximate size of uncompressed dataset: %.2f MB' % (
ds.nbytes / 1024**2
))
Approximate size of uncompressed dataset: 960.49 MB
Here we take a single time slice and try to figure out the best way to get a Beaufort-Only subset
time_slice = ds.isel(time=1200)
time_slice['age_of_sea_ice'].data
What does the slice look like?
time_slice['age_of_sea_ice'].plot(figsize=(11,11), add_colorbar=False);
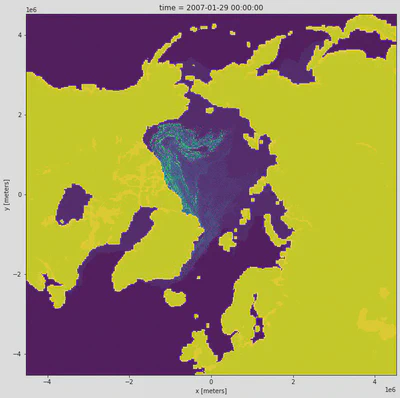
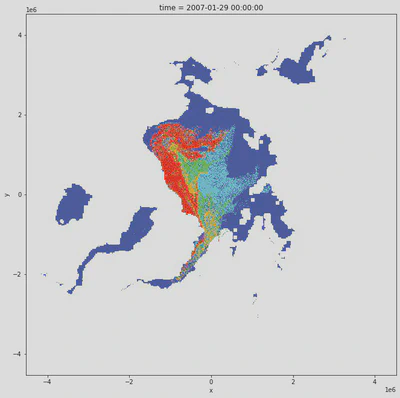
Let’s plot with the NSIDC colormap
# this implementation is imperfect but seems to work okay
cmap = ListedColormap(
[
'#FFFFFF', '#3F51A3', '#6FCBDB', '#69BF4E', '#F3B226',
'#F12411', '#F12411', '#F12411', '#F12411', '#F12411',
'#F12411', '#F12411', '#F12411', '#F12411', '#F12411',
'#F12411'
],
name='Sea Ice Age',
N=16,
)
cmap.set_over('#A1A1A1')
cmap
# note how we mask out the land/channels with a nice little conditional
xr.where(
time_slice['age_of_sea_ice'] >= 20,
np.nan,
time_slice['age_of_sea_ice']
).plot(
figsize=(11,11),
cmap=cmap,
add_colorbar=False
);
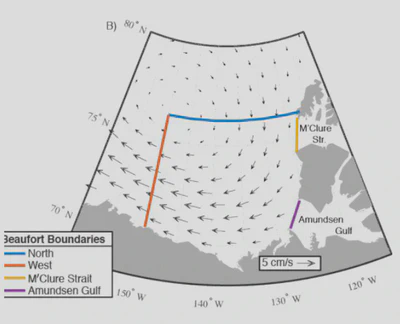
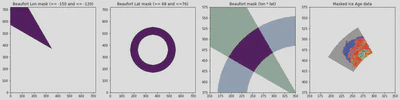
Create a Mask based on Dave’s Figure 1B: https://doi.org/10.1002/essoar.10509833.1
min_lon = -150
max_lon = -120
min_lat = 68
max_lat = 76
lons = time_slice['longitude'].data.copy()
lons[lons < min_lon] = np.nan
lons[lons > max_lon] = np.nan
lons[~np.isnan(lons)] = 1
lats = time_slice['latitude'].data.copy()
lats[lats > max_lat] = np.nan
lats[lats < min_lat] = np.nan
lats[~np.isnan(lats)] = 1
mask = lons * lats
fig, (
ax_lons, ax_lats, ax_mask, ax_data
) = plt.subplots(ncols=4, nrows=1, figsize=(18,6))
ax_lons.imshow(lons, origin='lower')
ax_lons.set_title('Beaufort Lon mask (>= -150 and <= -120)')
ax_lats.imshow(lats, origin='lower')
ax_lats.set_title('Beaufort Lat mask (>= 68 and <=76)')
ax_mask.imshow(mask, origin='lower')
ax_mask.imshow(lons, origin='lower', cmap='Greens_r', alpha=0.4, zorder=-2)
ax_mask.imshow(lats, origin='lower', cmap='Blues_r', alpha=0.4, zorder=-2)
ax_mask.set_title('Beaufort mask (lon * lat)')
ax_mask.set_xlim(150, 350)
ax_mask.set_ylim(375, 575)
ax_data.imshow(time_slice['age_of_sea_ice'] * mask, cmap=cmap, vmin=0, vmax=16)
ax_data.set_title('Masked Ice Age data')
ax_data.set_xlim(150, 350)
ax_data.set_ylim(375, 575)
fig.tight_layout();
With these two index ranges we can minimize our masked dataset
# just eyeballed these based on the above figure
xs = xr.DataArray(range(220, 300), dims='x')
ys = xr.DataArray(range(415, 535), dims='y')
(time_slice['age_of_sea_ice'] * mask).isel(
x=xs, y=ys
).plot(cmap=cmap, vmin=0, vmax=16, add_colorbar=False);
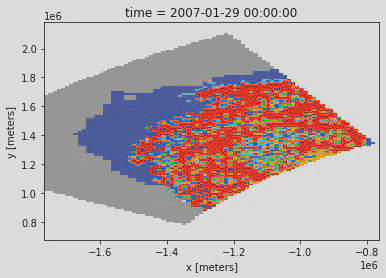
Now that we have our slicers we can reduce the dataset to just the Beaufort AOI
result = (ds['age_of_sea_ice'].isel(x=xs, y=ys) * mask[415:535, 220:300])
result
print('Approximate size of minimized dataset: %.2f MB' %
result.nbytes / 1024**2
))
Approximate size of minimized dataset: 70.46 MB
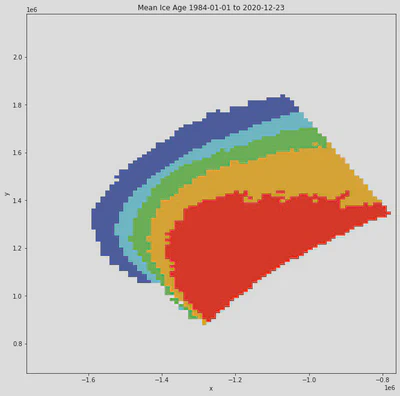
fig, ax = plt.subplots(figsize=(11,11))
xr.where(
result >= 20,
np.nan,
result
).mean(dim='time').plot(
cmap=cmap,
ax=ax,
vmin=0,
vmax=16,
add_colorbar=False
)
ax.set_title(
'Mean Ice Age ' +\
result["time"].min().dt.strftime("%Y-%m-%d").data +\
' to ' +\
result["time"].max().dt.strftime("%Y-%m-%d").data}
);
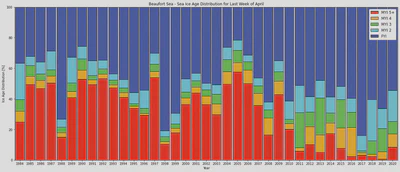
What about the last week of April like in Dave’s paper?
# any ice age greater than 5 will be given the "5+" moniker
data = xr.where(result >= 20, np.nan, result)
data = xr.where(data > 5, 5, data)
# imperfect but works
last_week_apr = data.loc[
(data['time'].dt.month == 4) &
((31 - data['time'].dt.day) < 5)
]
assert \
len(last_week_apr['time']) == \
len(range(
data['time.year'].min().values,
data['time.year'].max().values+1
))
last_week_apr
Now that we have our last-week-of-April timeseries we can construct a nice figure
df = pd.DataFrame({
'Year': [],
'FYI': [],
'MYI 2': [],
'MYI 3': [],
'MYI 4': [],
'MYI 5+': []
})
# if it's stupid and it works - it's not stupid
for year in last_week_apr['time.year'].values:
flat = last_week_apr.isel(time=last_week_apr['time.year'].isin([year])).values.ravel()
vals, counts = np.unique(flat[~np.isnan(flat)], return_counts=True)
if vals[0] != 0:
df = df.append(
pd.DataFrame({
'Year': [year],
'FYI': [counts[0]],
'MYI 2': [counts[1]],
'MYI 3': [counts[2]],
'MYI 4': [counts[3]],
'MYI 5+': [counts[4]]
}),
ignore_index=True
)
else:
df = df.append(
pd.DataFrame({
'Year': [year],
'FYI': [counts[1]],
'MYI 2': [counts[2]],
'MYI 3': [counts[3]],
'MYI 4': [counts[4]],
'MYI 5+': [counts[5]]
}),
ignore_index=True
)
df = df.set_index(df['Year'].astype('uint32')).drop('Year', axis=1)
# convert pixel counts into fraction of total
totals = df.sum(axis=1) / 100
for col in df.columns:
df[col] /= totals
fig, ax = plt.subplots(figsize=(22,9))
# need to invert column order to match dave's figure 3c
df[df.columns[::-1]].plot(
kind='bar',
stacked=True,
ax=ax,
width=0.9,
color=['#3F51A3', '#6FCBDB', '#69BF4E', '#F3B226', '#F12411'][::-1],
edgecolor='k'
)
ax.set_xlim(-0.5, len(df)-0.5)
ax.set_ylim(0, 100)
ax.set_ylabel('Ice Age Distribution [%]')
ax.legend(framealpha=1, loc='upper right', fontsize=12)
[lab.set_rotation(0) for lab in ax.get_xticklabels()]
ax.set_title('Beaufort Sea - Sea Ice Age Distribution for Last Week of April');
